DICOM Web-Viewer – VISILAB
A Web Viewer to explore microscopic images has been implemented by VISILAB. The viewer allows uploading many image formats including .svs, visualizing the image at different magnifications, creating annotations and selecting regions of interest. The size of microscopic images obtained from whole slide tissue samples are usually larger than 1GB, therefore it is impossible to visualize them with common software. The DICOM Viewer can work with such images, providing a useful tool for Digital Pathology. https://authors.elsevier.com/a/1ZPDqcV4L36NV
DICOM VISILAB WSI-Viewer See on YouTube
VISILAB WSI-Viewer See on YouTube
DICOM file converter – SAS – SESCAM
A library to convert all type and formats of whole slide images (WSI) into DICOM format to be used in digital pathology has been developed by SAS-SESCAM group. This converter has been demonstrated in the showcase called “DICOM Digital Pathology Connectathon”, that took place in San Diego, CA, in October, 2017. The converter was showed together with the above mentioned viewer. The entire system was successfully interoperable with several slide scanners and PACS systems from other manufacturers.
ANGIOPATH – VISILAB
ANGIOPATH is a morphometric tool allowing us to measure different aspects of the shape and size of vascular vessels in a complete and accurate way. The developed tool detect and close all vessels providing useful parameter for angiogenesis research. Therefore, ANGIOPATH is based on vessel closing which is an essential property to properly characterize the size and the shape of vascular and lymphatic vessels. The method is fast and accurate improving existing tools for angiogenesis analysis. ANGIOPATH also improves the accuracy of vascular density measurements, since the set of endothelial cells forming a vessel is considered as a single object.
http://dx.doi.org/10.1155/2013/263190
TMA_Core_Extraction – VISILAB
This tool allows automatic segmentation and archiving of tissue microarray (TMA) cores in microscopy images at different magnifications. A crucial step to improve the speed and quality of TMA analysis is the correct localization of each tissue core in the array. However, usually the tissue cores are not aligned in the microarray, the TMA cores are incomplete and the images are noisy and with distorted colors. VISILAB has developed a robust framework to handle core sections under these conditions. The algorithms are able to detect, stitch and archive the TMA cores at different magnifications. Once the TMA cores are segmented they are stored in a relational database allowing their processing for further studies of benign-malignant classification. The method was shown to be reliable for handling the TMA cores and therefore enabling further large scale molecular pathology research. http://dx.doi.org/10.1109/JBHI.2013.2282816
TMA_CAD_AIDPATH – VISILAB
A TMA Computer Aided Diagnosis (CAD) system has been implemented to provide support to pathologists in their daily work. For that purpose, the tool covers each and every process from the TMA core image acquisition to their individual classification. The classification process is based on texture properties and allows performing the core classification selecting different types of color models, texture descriptors and classifiers. The CAD system has been tested with breast TMA samples and it provides a classification of the TMA cores into three categories: malignant, doubtful and benign.
CROWDSOURCED IMMUNOHISTOCHEMISTRY
To experiment human computing in digital pathology images, UNIUD developed a crowsourcing application hosted by one major crowdsourcing site. The application can be used to collect a number of human evaluations of immunohistochemical nuclear markers and aggregate them to obtain a possibly reliable result, according to crowdsourcing principles. The application has been successfully tested on MIB1 stained breast cancer images, with very high correlation vs. expert evaluation (doi: 10.1186/1746-1596-9-S1-S6). While unlikely usable in clinical routine, crowdsourcing might provide a fast lane to image evaluation in research contexts.
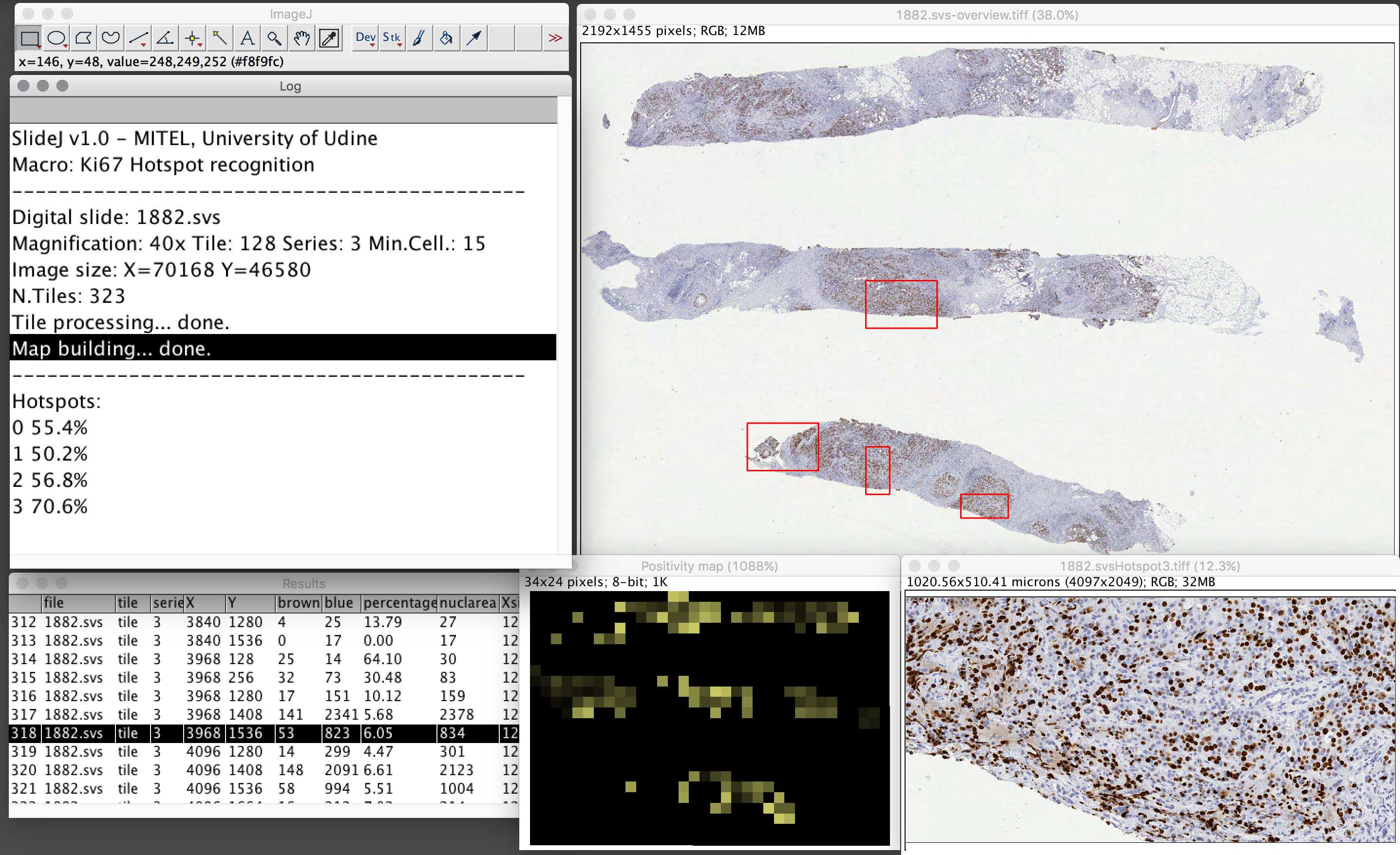
The SlideJ plugin for processing digital slides on ImageJ
The digital slide, or Whole Slide Image, is a digital image, acquired with specific scanners, that represents a complete tissue sample or cytological specimen at microscopic level. While Whole Slide image analysis is recognized among the most interesting opportunities, the typical size of such images—up to Gpixels- can be very demanding in terms of memory requirements. Thus, while algorithms and tools for processing and analysis of single microscopic field images are available, Whole Slide images size makes the direct use of such tools prohibitive or impossible. In this work a plugin for ImageJ, named SlideJ, is proposed with the objective to seamlessly extend the application of image analysis algorithms implemented in ImageJ for single microscopic field images to a whole digital slide analysis. The plugin has been complemented by examples of macro in the ImageJ scripting language to demonstrate its use in concrete situations. https://doi.org/10.1371/journal.pone.0180540
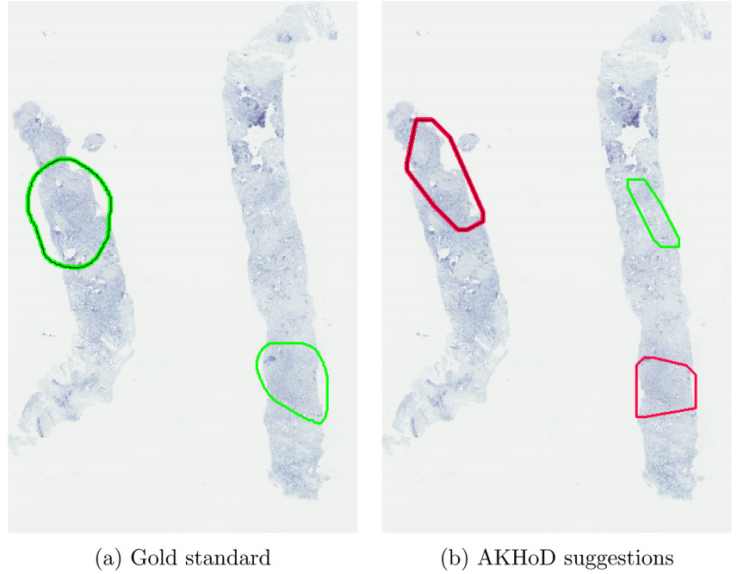
AKHOD – UNIUD
A software for detecting Ki67 hotspots on digital slides, called AKHOD, has been developed and tested on 50 biopsies, with behaviour slightly better than a single pathologist. Details of this tool may be found at: ‘An adaptive positivity thresholding method for automated Ki67 hotspot detection (AKHoD) in breast cancer biopsies’. Comput Med Imaging Graph (2017), http://dx.doi.org/10.1016/j.compmedimag.2017.04.005.